Fusion Model
Overview
The Early Fusion Model integrates diverse data types using separate architectures optimized for each modality and combines them into a final model for overall prediction. This holistic approach is particularly valuable for our project, where integrating structured data, text, and images can provide a comprehensive understanding of patient conditions.
Model Workflow
The model workflow can be summarized as follows:
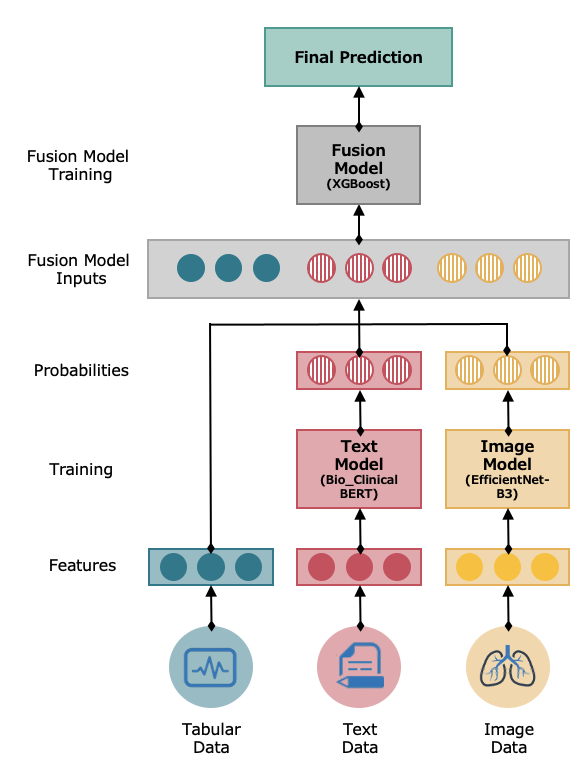
Data Inputs
The model takes the following inputs:
- Tabular Data: Structured data with rows and columns, including patient’s data and vital signs.
- Text Data: Unstructured text data from clinical notes.
- Image Data: Imaging data in the form of chest x-rays.
Individual Model Training
Both the text and image data are processed by a model that is best suited for that particular type.
- Text Model: The text model is an NLP-based multiclass classification system designed specifically for medical text analysis. Its basis is a pre-trained BERT model called Bio ClinicalBERT (opens in a new tab), which was trained on all MIMIC-III notes and initialized from BioBERT (opens in a new tab).
- Image Model: The image model is a scaling method for convolutional neural networks (CNNs) that uniformly scales all dimensions of depth/width/resolution. For our model, we used EfficientNet-B3, which is a specific version of EfficientNet that has been pre-trained on a large dataset of images.
Fusion Model Inputs
The outputs of the text and image models are a set of probabilities or a feature vector (5 features) that indicates its predictions. In the context of classification, these probabilities would represent the likelihood of each class being the correct one. These outputs and the original tabular data (8 features) are then combined (concatenated) to serve as inputs for the fusion model (total of 18 features).
Fusion Model Training
-
Data Preparation
- The fusion model leverages 18 features compiled from the individual models' outputs and the tabular data. The target for these inputs is a multi-class diagnosis, categorized into one of five classes.
-
Choosing a Base Model
- A basic XGBoost model is selected for its effectiveness in handling diverse datasets and its robustness in classification tasks.
-
Cross-Validation Methodology
- K-Fold Cross-Validation: To ensure the model's robustness and generalizability, k-fold cross-validation is implemented. This technique enhances validation accuracy and reduces the risk of model overfitting.
-
Hyperparameter Tuning
- Through systematic grid search, multiple combinations of XGBoost parameters (e.g., learning rate, max depth, and number of trees) are tested to identify the optimal settings.
-
Performance Evaluation
- The model's effectiveness is measured using Negative Log Loss and AUC metrics. These metrics assess the model’s ability to accurately classify and differentiate between the diagnosis classes.
-
Model Selection and Final Testing
- The final model is chosen based on the best average performance across all cross-validation folds.
- An independent test set is used to validate the model's real-world applicability and to assess its generalization capability.
Fusion Model Prediction
-
Input Data Preparation
- When new data is received, it is prepared and formatted to ensure it aligns with the model’s requirements, maintaining the structure of the 18-feature input.
-
Model Loading
- The optimally tuned XGBoost model, trained with data from the fusion input, is loaded for prediction tasks.
-
Making Predictions
- The model processes the input features to predict the likelihood of each diagnosis class. The final output is a tensor of dimensions (1, 5), representing the confidence in each class.
-
Interpretation and Use
- The probabilities are used to determine the most likely diagnosis, aiding healthcare professionals in decision-making processes.